1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
|
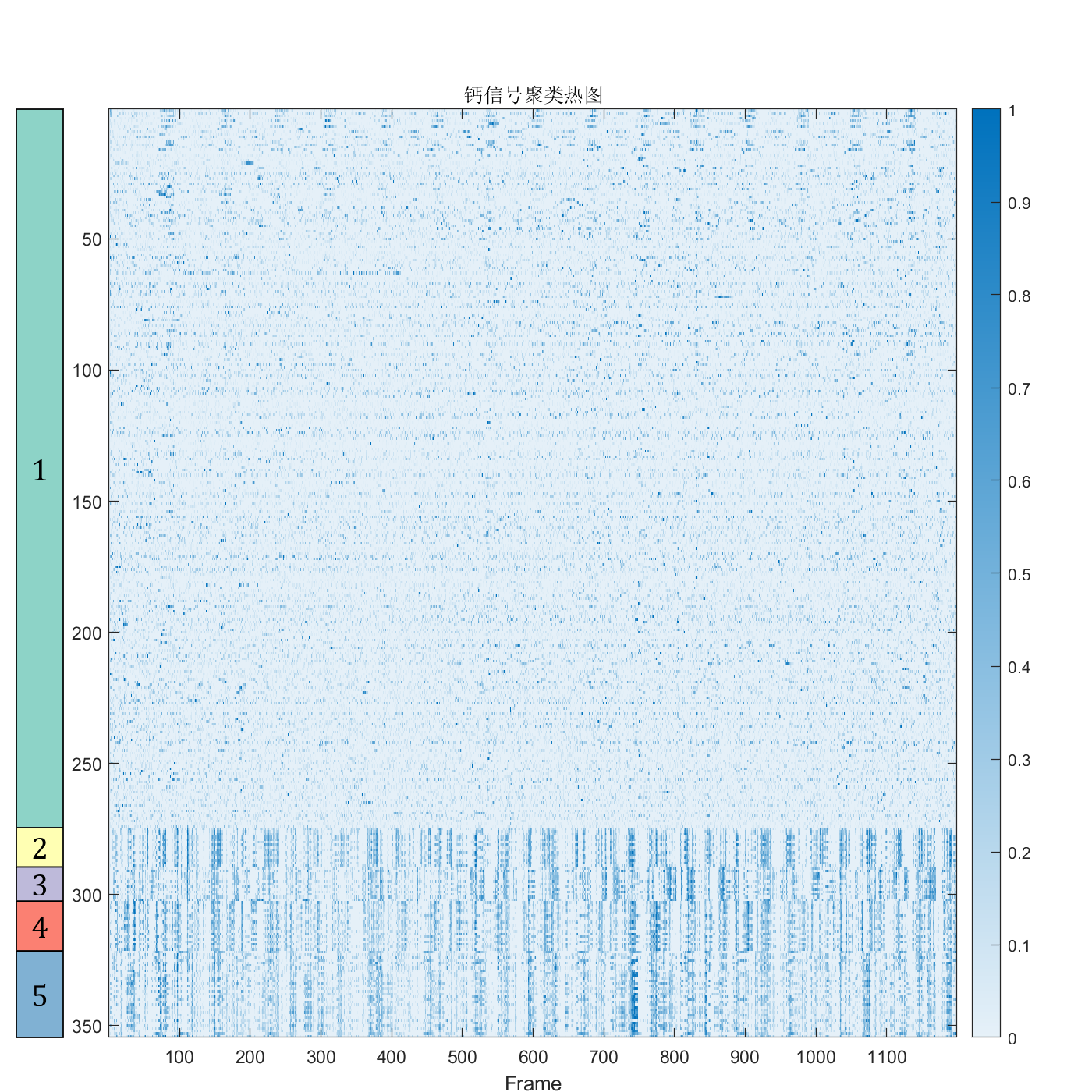
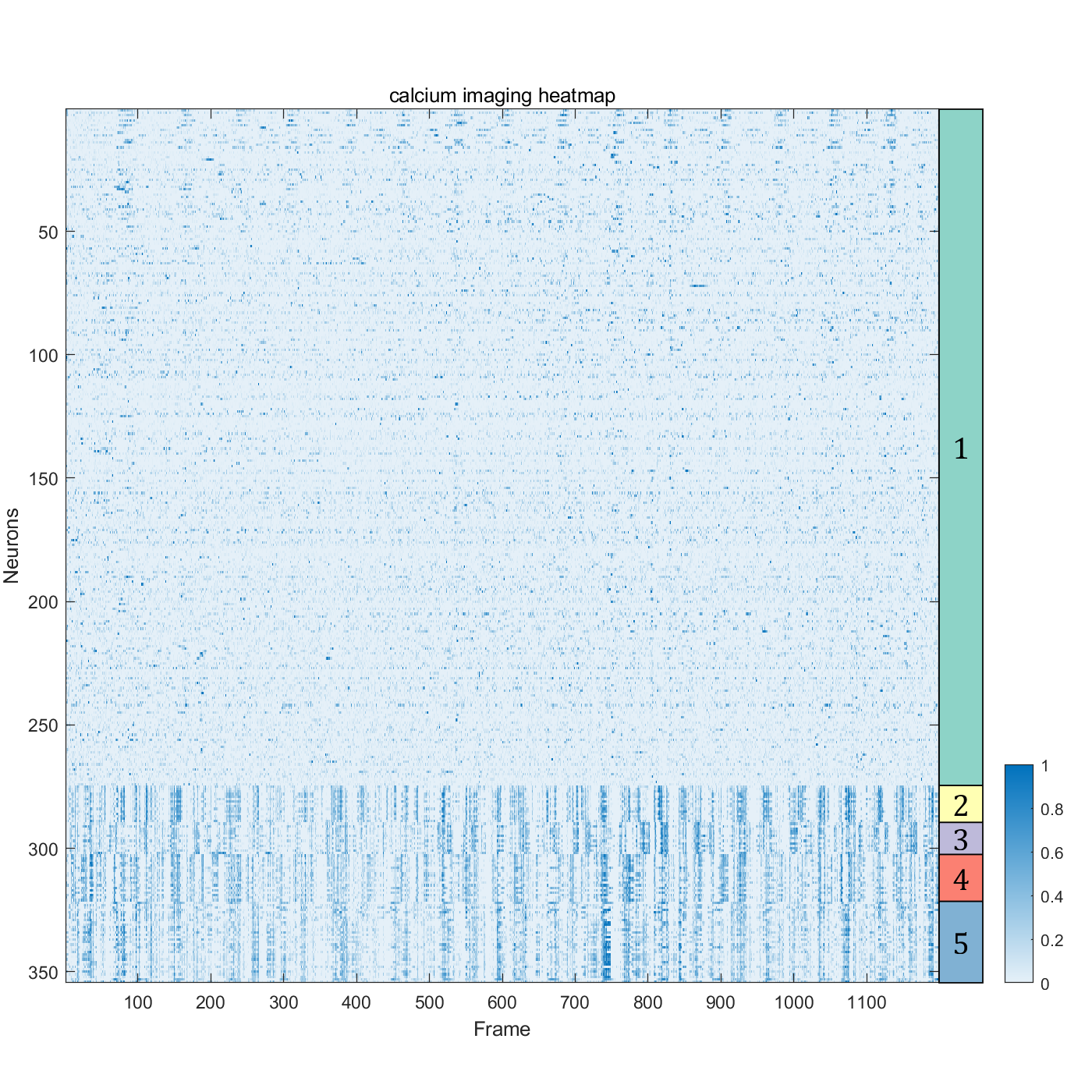
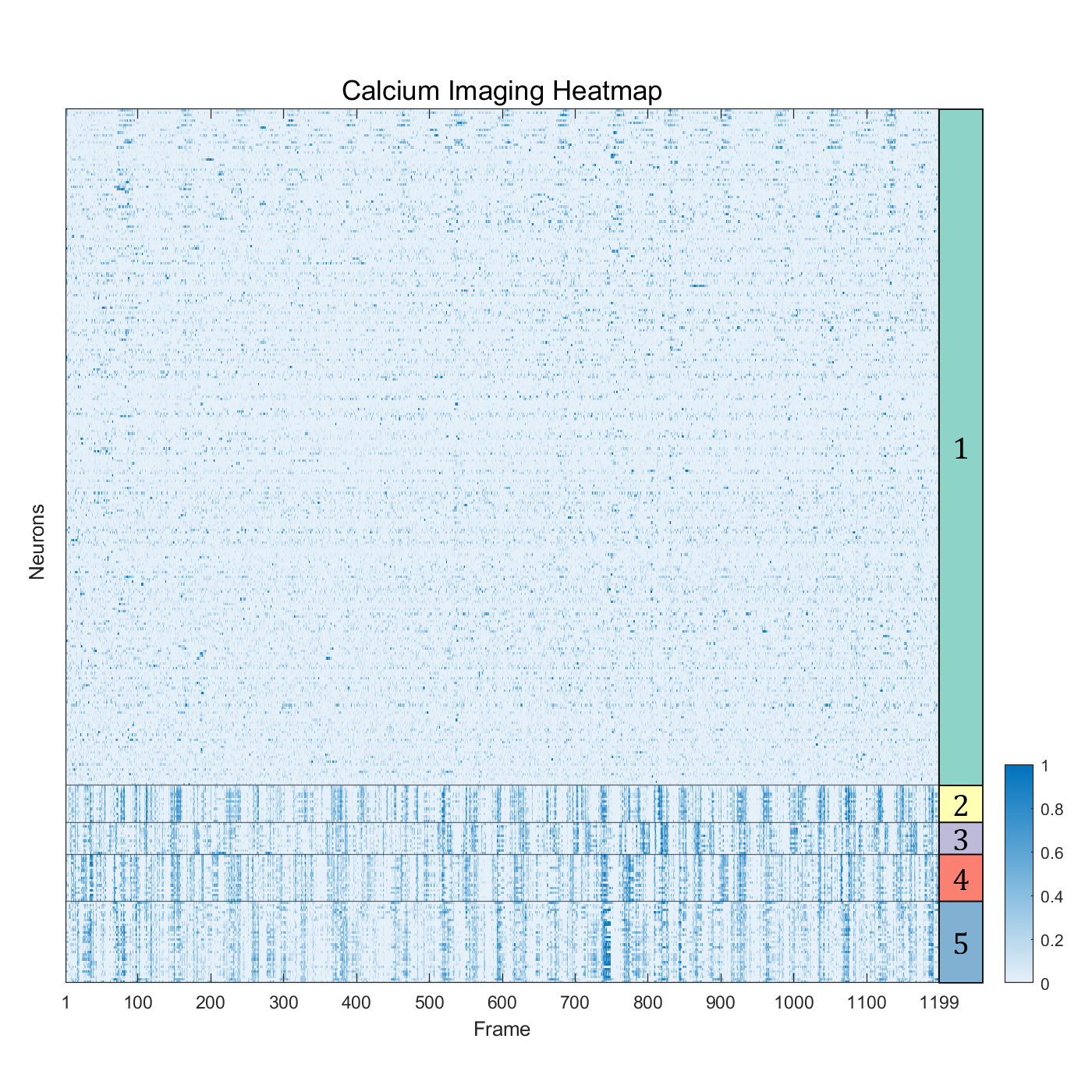
data = readtable('suite2p_ROI_data_added_columns.csv','ReadVariableNames',true)
cells = data(data.is_cell == 1,:)
cells_activity = cells(:, 2:1200)
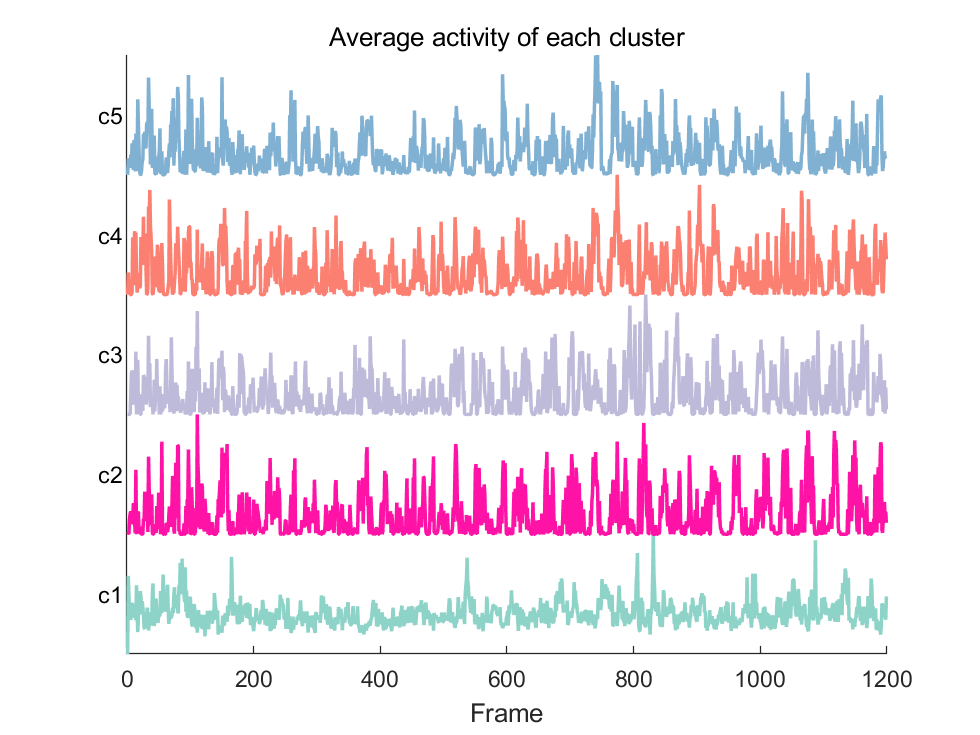
numClusters = 5;
data_t = table2array(cells_activity);
[idx, centroids] = kmeans(data_t, numClusters,'Start', ones(5,1199));
class_name = string(1:numClusters);
[sortedResult, sortedIdx] = sort(idx);
sortedNeuronData = data_t(sortedIdx, :);
fig=figure('Position',[100,100,800,800]);
axMain=axes('Parent',fig);
axMain.Position=[0.06,.1,.80,.80];
P=axMain.Position;
imagesc(axMain,sortedNeuronData);
axMain.YAxisLocation='left';
colormap(sky);
cb = colorbar('Location','eastoutside');
set(cb,"Position",cb.Position+[0.12,0,0,-0.6]);
xlabel('Frame');
ylabel('Neurons');
title('calcium imaging heatmap');
axBlockL=axes('Parent',fig);
axBlockL.Position=[0.86,P(2),P(3)/20,P(4)];
[X,Y]=SClusterBlock(sortedResult,'Orientation','left','Parent',axBlockL);
for i=1:length(X)
text(axBlockL,X(i),Y(i),class_name(i),'FontSize',17,'HorizontalAlignment','center','FontName','Cambria')
end
|